Wastewater monitoring dashboard: Technical notes
This dashboard provides data about COVID-19, flu, RSV and mpox virus levels in wastewater.
- Last updated: 2024-11-12
This page has information about how we conduct wastewater testing and the limitations of the data. It also includes definitions for some of the scientific terms used in this dashboard.
On this page
Methodology
Scientists across the country provide wastewater monitoring data through their provincial and territorial networks. To detect viruses at the community or institutional level, samples are taken at a central collection point, such as wastewater treatment plants or pumping stations. This method only captures the presence of viruses in the community or institution. It can’t be used to identify single cases or households.
Scientists continue to improve methods for detecting and measuring COVID-19, flu, RSV and mpox in wastewater. The scientific community, including the Public Health Agency of Canada, is working together to build a standard that will help everyone understand, compare and share data about respiratory viruses and mpox virus in wastewater. The results shown on this page were obtained by PCR testing and genomic sequencing.
Scientists use genomic sequencing to decipher the different genetic fragments of the virus found in the wastewater samples. Once the sequencing reaction is complete, they analyze the sequenced pieces using special software. These programs provide information on the variants and the relative amount of each variant detected in a wastewater sample.
We’ve compared wastewater signal trends when the same sites are tested by both the National Microbiology Laboratory (NML) and provincial and territorial networks. We found that the trends are broadly consistent across labs. Differences in the strength of the wastewater signals are mostly due to differences in processing methods.
We present COVID-19, flu and RSV wastewater viral load testing as a population-weighted 7-day rolling average. This is because high levels on a single day don’t show the broader trend. Our approach helps us to understand the overall trends while giving you better information to make your own health decisions. Generally, we test sites twice weekly. Exceptions are Alberton and Winnipeg which are tested 1 and 5 times per week, respectively.
How we calculate the EpiWeek signal for respiratory viruses
We calculate the EpiWeek signal by averaging all wastewater viral signals from that EpiWeek. Wastewater signals are aggregated to the EpiWeek for all collection sites within the same region as these sites can have different sampling schedules.
As new data becomes available, we may update the signals of the latest reported EpiWeek.
How we calculate the wastewater activity level index for respiratory viruses
To provide more context to COVID-19, flu and RSV signals, we’ve developed an activity level index. It’s based on a combination of the signal level and signal trend metrics. We need at least 12 months of data, excluding non-detects, to calculate the signal level and trend metrics.
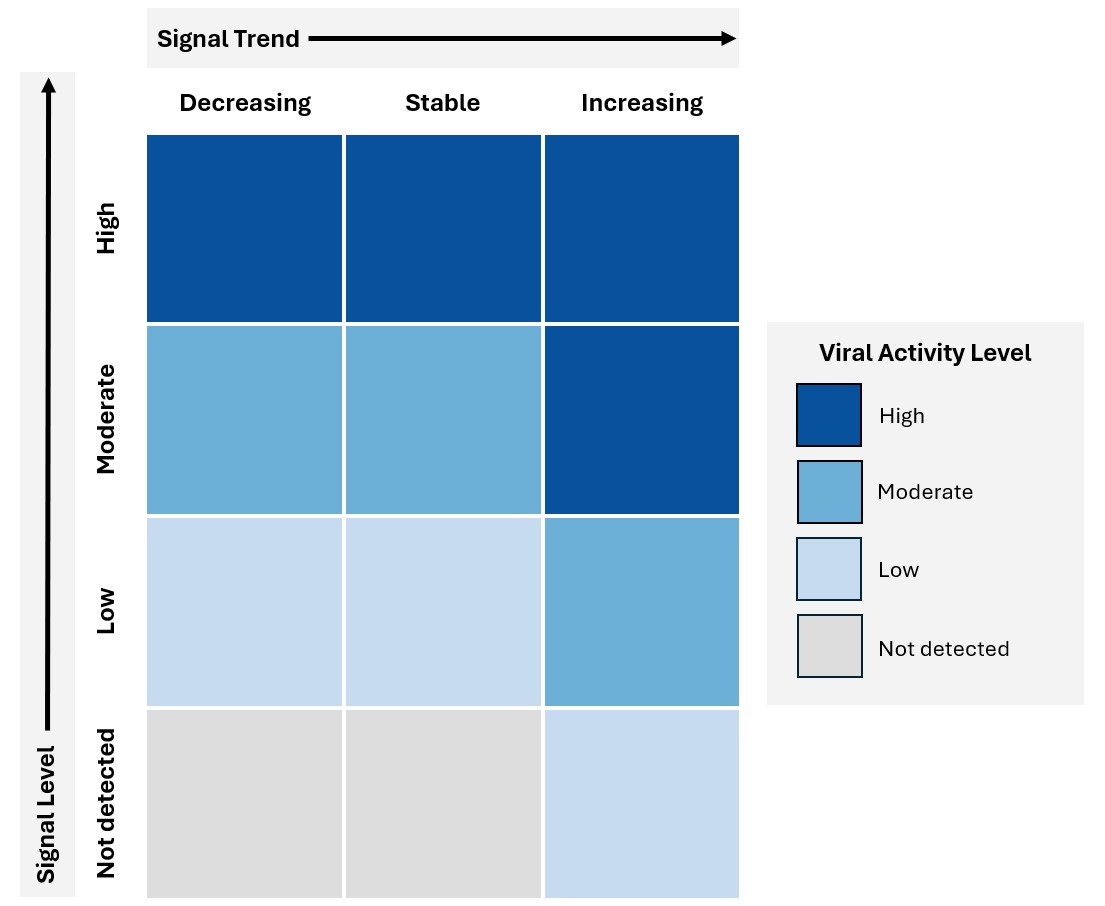
- High activity level means:
- viral signals are high compared to values over the past year, or
- viral signals are moderate compared to values over the past year, and the recent trend is increasing
- Moderate activity level means:
- viral signals are moderate compared to values over the past year, and the recent trend is decreasing or stable, or
- viral signals are low compared to values over the past year and the recent trend is increasing
- Low activity level means:
- viral signals are low compared to values over the past year, and the recent trend is decreasing or stable, or
- viral signals are below the threshold for detection, and the recent trend is increasing
- No detection activity level means:
- viral signals are below the threshold for detection and the recent trend is decreasing or stable
Calculation of signal level metric
The signal level metric describes the most recent COVID-19, flu and RSV viral loads compared to values over the past year. For each sampling location, we use viral loads from all samples collected 1 year before the most recent sampling period, to:
- calculate the 25th and 75th percentiles
- establish lower and upper cutoff values
Values below the 25th percentile are classified as low and values above the 75th percentile are classified as high. The rest are classified as moderate. Sites are marked as new if there are less than 12 months of data, excluding non-detects.
Calculation of signal trend metric
We monitor the rise and fall of COVID-19, flu and RSV signals using a technique developed by the Ontario government. The wastewater monitoring 7-day average data is broken into segments over time. The daily change in the viral signal is determined for each segment. Rises and falls of the wastewater signal are judged based on their consistency over time.
For more information, please refer to:
The signal trend metric describes how wastewater viral loads are changing based on the previous 5 weeks:
- Increasing is a statistically significant increase in the wastewater signal.
- Stable is either a steady signal or an insignificant decrease in the wastewater signal.
- Decreasing is a statistically significant decrease in the wastewater signal.
How we conduct geographical aggregation for respiratory viruses
Calculation of viral load
Geographical aggregation to city, province, territory or nation is the average of all collection sites within the respective area. The average is adjusted for population coverage. This means that collection sites with higher populations have a larger impact on the overall value. The greater the population coverage, the more representative the wastewater signal is of the overall community disease burden. Higher population coverage also increases the probability of detecting local outbreaks. Sites with low population coverage (less than 25% of the population) are indicated below the graphs.
In each graph, the grey shaded area shows the minimum and maximum wastewater signal of each EpiWeek since the beginning of the surveillance period. Geographical aggregates of viral load are only reported when data is available for at least half of the reported population is not available. At the national level, we only report aggregated data when data is available for at least 75% of the reported population.
Calculation of activity level index
- High = 3
- Moderate = 2
- Low = 1
- Not detected = 0
Geographical aggregation of the activity level to city, province, territory or nation is the average activity level of all collection sites within the respective area. The average is adjusted for population coverage. The population-weighted average is assigned an activity level according to the following criteria:
- 0 = Not detected
- 1.5 – 0 = Low
- 2.5 – 1.5 = Moderate
- 3 – 2.5 = High
How mpox testing and analysis is conducted
Mpox virus is found in wastewater in very low quantities, between 10 and 100 times lower than respiratory viruses. Consequently there is a high uncertainty in any single measure of the virus in wastewater. Therefore, we interpret signals across multiple EpiWeeks to reduce this uncertainty.
The NML tests for the presence and abundance of mpox virus in wastewater through 2 independent laboratory tests. A general mpox test is sensitive to all clades of mpox: for each sample, 6 replicates are tested across 2 different tests. Our map and dashboard reflects testing for general mpox and signals do not represent any specific clade. Additionally, a single clade I specific test is tested across 9 replicates per sample. A confirmed clade I detection will be communicated in text on the main results page of the dashboard.
The detection of mpox in wastewater is classified as:
- Consistent detection indicates that the mpox virus was detected in the wastewater for more than 2 weeks in the past 4 weeks.
- Intermittent detection indicates that the mpox virus was detected for 2 weeks in the past 4 weeks.
- No detection indicates that the mpox virus was not detected or detected for only 1 week in the past 4 weeks.
- No recent data indicates that less than 2 weeks of data was gathered to make an assessment about the presence of mpox virus in the wastewater in the past 4 weeks. This is usually due to delays in receiving samples.
Limitations
While wastewater monitoring offers many advantages, it does have some limitations.
The accuracy of the wastewater signal can be affected by various factors, including the composition of wastewater, which varies by community. For example, ground or surface water can make the wastewater signal stronger or weaker. This can be an issue during seasonal snow-melt and large rain events.
The wastewater signal can also be affected by:
- industrial flow into the wastewater collection system
- sand and salt to roads in winter
- the temperature of the collection system
- the method used for collecting the sample
- the diversity of variants in a sample
- the presence of similar mutations found in different variants
- changes in precipitation, including snowmelt
We’re working with our partners to identify other issues with wastewater monitoring and developing measures to reduce the effects.
Considering the above limitations, we’re not sure how much virus is shed with each wave. For this reason we don’t recommend comparing wastewater monitoring data from different waves of respiratory viruses to estimate the number of cases in a community.
Definitions
- The 7-day average is generated by averaging the levels from a given day with the 6 previous days. The average is termed “rolling” as it changes each day.
- Copies per mL is the number of copies of the target RNA found in a milliliter (mL) of raw wastewater by the specific wastewater treatment facility.
- Viral load is the concentration of SARS-CoV-2, Influenza, or RSV genetic material present in a sample of wastewater. We present this in the dashboard as copies of SARS-CoV-2, Influenza, or RSV genetic material per milliliter (mL) of wastewater.
- Wastewater signal is a measure of the level of virus in wastewater that identifies increasing, stable or decreasing number of virus particles in wastewater.
- 25th percentile is also called the first quartile. 25% of the data falls below this level.
- 75th percentile is also called the third quartile. 75% of the data falls below this level.
- Non-detect is when the detection limit of the respiratory virus was not reached during testing of the wastewater sample.
- Epidemiological week (EpiWeek) is an approach to counting weeks that allow for comparison of data between years. For the EpiWeek appearing on the x-axis of the graphs we have used Sunday as the first day of the week.
To learn more about wastewater monitoring, please refer to:
Data changes
| Date | Notes |
|---|---|
| 2023-05-02 | We have temporarily removed Saint John from the dashboard, due to possible issues affecting data accuracy. Once these issues have been investigated and resolved, Saint John will once again be included in the dashboard. |
| 2023-05-12 | In January 2023, The NML updated its protocol to include quantification of standard reference samples to improve accuracy. This change affects data points between July 6, 2022 and February 3, 2023. Data points have been retroactively updated to reflect this change. The updated NML wastewater quantification protocol includes a confirmation of standard reference samples’ concentration via digital PCR. |
| 2023-09-08 | Saint John, New Brunswick, has been reincluded in the dashboard with data originating from Dr. Georges-L.-Dumont University Laboratory. |
| 2023-12-15 | Surveillance data for Bathurst, Campbellton, Fredericton, Miramichi and Moncton has been replaced by data originating from Vitalité Health Network in New Brunswick. Historical data analyzed by the NML is still available for download. |
| 2023-12-15 | Historical data for Edmundston and Saint John have been updated with a new quantification method to more accurately reflect the laboratory process. |
| 2024-02-23 | Wastewater activity updates for the following Saskatchewan sites are on hold due to a pause in samples sent to PHAC:
|
| 2024-07-02 | The inclusion of data from Prince Edward Island is currently paused. Following approval, data from Prince Edward Island will once again be included in the dashboard. |
| 2024-07-19 | Prince Edward Island data has been reincluded in the dashboard following provincial review. |
| 2024-08-20 | The inclusion of new data from New Brunswick is currently paused while a quality review of the data is being conducted by the NML. |
| 2024-09-06 | Data from the Toronto North wastewater site for EpiWeek 34 has been removed from the weekly updates. The data from this week was unusual and is unlikely to be linked to an epidemiological event but we are further investigating this signal. |
| 2024-09-20 | New Brunswick data has been reincluded in the dashboard following the quality review. |
| 2024-12-09 | Data from a Central Colchester wastewater sample collected on EpiWeek 41 has been removed from the weekly updates. The data from this week was unusual and is unlikely to be linked to an epidemiological event but we are further investigating this signal. |
- Date modified: